Cancer Evolution
Recent progression of cancer genome projects has uncovered the mutational landscapes of many cancers, but how cancer cell evolves with and without therapy is still unclear. Scientists believe one major reason of treatment failure is the temporal-spatial dynamics of cancer cells. Actually, cancer cells are constantly evolving, with different groups of cells accumulating distinctive mutations. As the search for more effective cancer diagnostics and therapies continues, remained key questions include a) how to interpret intratumor heterogeneity (ITH); b) how to understand the tumors change over time and how to predict the impact of ITH on tumor progression; and c) how to disentangle the order in which mutations occur. Being able to predict how a tumor will behave based on signs seen early in the course of disease could enable the development of new diagnostics that could better inform treatment planning.
Precision Medicine
Traditional cancer treatment includes surgery,chemotherapy and radiation therapy. Different from conventional chemotherapy that tends to destroy all rapidly dividing cells, targeted therapy aims at cells characterized by particular markers. For example, Imatinib targets BCR-ABL positive chronic myeloid leukemia; Crizotinib targets Alk-fussion lung cancer; Vemurafenib targets B-Raf mutant melanoma, etc. Although optimally managed, many cancers might relapse, and scientists believe the treatment failure is related to both inter- and intra- genomic/phenotypic heterogeneity of cancer cells. To reveal the complexity of cancer development and progression, and to identify novel targetable cancer alterations for personalized medicine, we will aim to (1) mine Large Cohorts of sequencing data to Identify Novel Driving Events; (2) Relate Personal Genomes to Tailored Treatment using machine learning methods.
Noncoding Genome
The human genome project has shown that only a small fraction (<2%) of human genome can be transcribed into mRNA that is further translated into protein, and the vast majority of the mammalian genome might express non-coding RNA (ncRNA). Although a number of long non-coding RNAs (lncRNAs) have been recently shown to play significant roles in the regulation of gene expression or protein activity in critical signaling pathways, the total number of ncRNAs and the fraction of functional ncRNAs within the mammalian genome are still mysteries. We are constructing computational algorithms and pipelines to assemble and quantify lncRNAs from transcriptomic data and elucidate their roles in cancer evolution, drug resistance, chromatin organization and genomic instability.
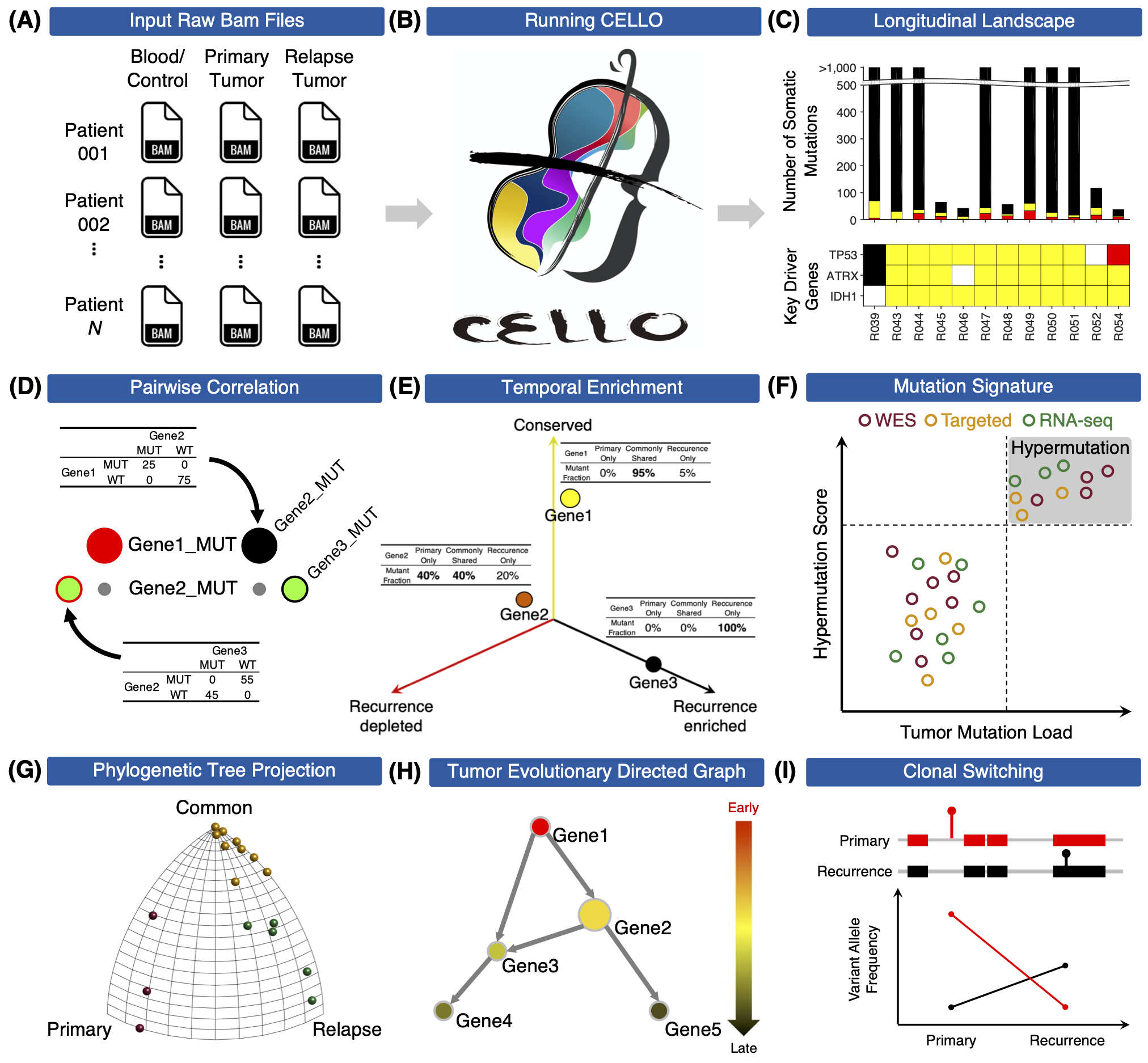
Tools for untangling cancer evolution from longitudinal genomic data
Targeting tumor-specific mutations via customized chemical compounds can precisely eradicate the cancer cells without harming healthy tissues, which paves a way toward precision oncology. But this precision oncology strategy has not been successful in many refractory cancers such as glioblastoma (GBM). One of the main obstacles is the limited understanding of cancer evolution, in which cancer cells might acquire advantageous fitness to revive under treatment stress.
To understand how cancer evolves under treatment stress, the Wang Lab developed CELLO (Cancer EvoLution for LOngitudinal data), to analyze and visualize longitudinal next-generation sequencing data before and after treatment. Particularly, CELLO can conduct the following analytical workflow including (1) generation of longitudinal mutational landscape, (2) detection of mutational signature for cross-platform sequencing data, (3) clustering of patients based on evolutionary patterns, (4) identification of clonal switching events; and (5) inference temporal order of somatic mutations. To benefit researchers who are interested in longitudinal cancer genomics study for analyzing their own data, both MATLAB and R versions of CELLO are developed. To ensure reproducibility and usability, we also present a docker version of CELLO based on the R implementation.
CELLO: a longitudinal data analysis toolbox untangling cancer evolution
Biaobin Jiang*, Dong Song*, Quanhua Mu, Jiguang Wang#
Quantitative Biology, in press, 2020
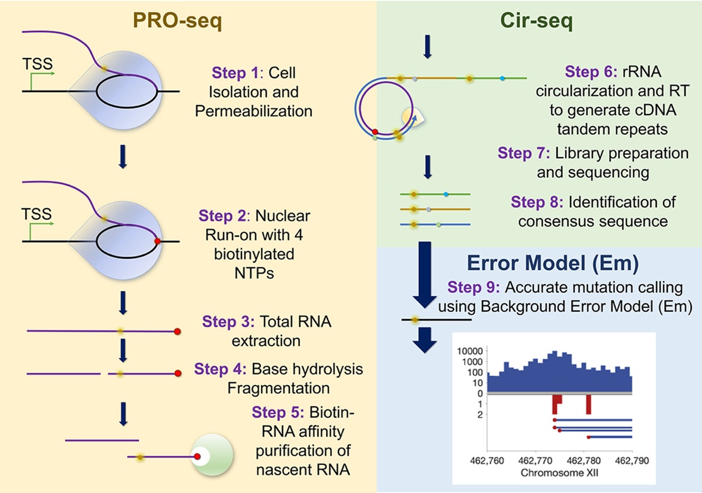
New sequencing protocol and model identify transcription errors
RNA polymerase transcribes certain genomic loci with higher errors rates. These transcription error-enriched genomic loci (TEELs) have implications in disease. Current deep-sequencing methods cannot distinguish TEELs from post-transcriptional modifications, stochastic transcription errors, and technical noise, impeding efforts to elucidate the mechanisms linking TEELs to disease.
Collaborating with Prof Xuhui Huang in HKUST Chemistry, we together describe background error model-coupled precision nuclear run-on circular-sequencing (EmPC-seq) to discern genomic regions enriched for transcription misincorporations. Applying EmPC-seq to the ribosomal RNA transcriptome, we show that TEELs of RNA polymerase I are not randomly distributed but clustered together, with higher error frequencies at nascent transcript 3′ ends. Our study establishes a reliable method of identifying TEELs with nucleotide precision, which can help elucidate their molecular origins.
Identifying Transcription Error-Enriched Genomic Loci Using Nuclear Run-On Circular-Sequencing Coupled with Background Error Modeling
Cheung, P.*, Jiang, B.*, Booth, G.T., Chong, T.H., Unartar, I.C., Wang, Y., Suarez, G.D., Wang, J.#, Lis, J.T.#, Huang, X.#.
J. Mol. Biol., 432(13):3933-3949, 12 June 2020